Antony N. Davies,a,b Peter Lampenc and Robert Lancashired
aExpert Capability Group – Measurement and Analytical Science, Nouryon, Deventer, the Netherlands
bSERC, Sustainable Environment Research Centre, Faculty of Computing, Engineering and Science, University of South Wales, UK
cLeibniz-Institut für Analytische Wissenschaften – ISAS, Dortmund, Germany
dThe Department of Chemistry, The University of the West Indies, Mona, Kgn 7, Jamaica
DOI: https://doi.org/10.1255/sew.2019.a1
As spectroscopists we tend to focus on spectra. Nothing new there then, but maybe we need to be paying more attention to the information surrounding our measurements which define the context and relevance of the data and in many cases the fundamental ability to display the spectra correctly. I must admit to having somewhat neglected the creation and fate of metadata in all the data handling and migration work we have carried out over the years. It was occasionally funny to see certain spectrometer manufacturers using the ##OWNER= field in JCAMP-DX files to claim they owned the spectra we were creating and to be honest we pretty much ignored this when helping the vendors to get the actual data migrations compliant to the standards. However, there are increasing demands on scientists to upload accompanying data with their peer-reviewed papers, companies to make better use of the big data and machine learning tools are becoming ever more accessible. This means it is the metadata which gives meaning to the measured spectra and it is the metadata which will probably outlive the original creator of the spectra and almost certainly the organisations within which the data were created. In a world where digital rights management is ever more important, do you really want to leave the ownership of your data in the hands of your instrument vendor?
Why are metadata so important?
Last year we reported from the IUPAC/CODATA workshop in Amsterdam “Supporting FAIR Exchange of Chemical Data through Standards Development”.1 This was followed up by IUPAC formally endorsing the Manifesto of the Chemistry GO FAIR Implementation Network (ChIN) on 28 January 2019. This chemistry network is part of a larger global science network that supports the FAIR guiding principles for scientific data management and stewardship.
In the original report last year, we mention that the follow-up would include a Project Group 2: focussing on metadata for data publication and the items that could be considered important to FAIRify the data. A workshop was held titled Fair Publishing Guidelines for Spectral Data and Chemical Structures on 29–30 March 2019 during the American Chemical Society National Meeting & Exposition in Orlando, Florida which threw up some interesting challenges for IUPAC and the future data standards work. The original JCAMP-DX formats were never designed to transport all the metadata from an instrumental measurement to a vendor-neutral file format. The JCAMP-DX CORE fields were just those essential to correctly reading and interpreting the data and which were accepted by all the vendors working on the development of the various standards. There are also many potential labels defined in the standards which were not essential, but their use was not controlled by us. We know of at least one vendor who developed a JCAMP-DX export software which made extensive use of the $$ prefixed private labels to export all their instrumental parameters to their JCAMP-DX files. This allowed the vendor to completely re-create the data set on another software system. As they were un-documented to the outside world could not be used by anyone except the vendor.
From my regulatory compliance experience any records created in a regulated environment fall under some record retention policy or other, so discussing whether to store individual bits of the record—the metadata—as if it had a life all on its own seemed utterly pointless. Indeed, the original FDA 21CFR part 11 guidelines created all sorts of questions about the use of the JCAMP-DX standard file format in this scenario as it did not require the storage of all the original metadata (see below for the new Guidance which makes it much easier to accept JCAMP-DX files in this environment).
As the discussions continued since last year, it has become clear that increasing demands on scientists publishing research to upload their “raw data” to some open public repository or other has caused issues when the only metadata available were linked to the publication rather than the data itself. This is fine if your scope is simply limited to locating data in the repository from the perspective of the specific publication they are cited in, but what if you would like to find all the 13C-NMR spectra measured with instruments with 500 MHz field strengths or better using deuterated chloroform as the solvent?
Metadata are critical to the correct functioning of our data systems!
My overly cited quote from Sherlock Holmes that “It is a capital mistake to theorise before one has data” from A Study in Scarlet has unfortunately turned on me and should now probably read “It is a capital mistake to theorise before one has data, and the associated domain-specific metadata to ensure that the data are Findable, Accessible, Interoperable and Reusable”.1,3,4
What do we need to consider here? I have recently talked to a system owner with a widely deployed Chromatography Data System (CDS) from one of the top international vendors. Even though the CDS was professionally deployed, maintained and continually updated to the latest release versions, the highly professional outsourced data storage provider had, without reference back to their customer, decided at some point into their contract not to back up the metadata tables in the database. I think you all know what is coming, yes… I will not spell it out as it is too painful but the inevitable did happen…. Major lesson learned (hopefully) to test your disaster recovery position REGULARLY! All the data could be restored but none of the metadata. Fortunately, as far as I know this system did not fall under any sort or regulatory compliance position. And to think of all the times I have jealously praised the chromatographers for having better and more reliable tools at their disposal than us poor spectroscopists!
Metadata is often described as information about data. The loss of metadata goes to show that metadata is critical for the operation of our scientific society in the short to medium term but in the longer term the metadata may well need to evolve as the context changes to remain relevant. In the short term, being able to identify five chromatograms, six NMR spectra and a couple of infrared spectra as being measured as part of a specific analytical question is essential to generating and validating results. In the longer term, these data sets may become part of a much greater set of essential evidence in proving a new drug is safe to use. After an audit the same data sets and the way they were processed could be called upon forming the basis of proving compliance to good laboratory practises for an organisation. In this way the “information about the data” and the way it is used can evolve.
The original Dublin Core
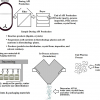
So, let us go back a little in history and look at one of the early initiatives to standardise on specific metadata fields to create some order out of the chaos of retrieving information from diverse sources and location in the internet age. Figure 1 shows an early attempt to start to define metadata than could be applied to any digital or physical object such as videos, pictures, web pages, books, DVDs, artworks or even spectra, known as the Dublin Core.4 Unfortunately for our Irish readers, the Core was named after the original invitational Metadata workshop called by The Online Computer Library Center (OCLC) and the National Center for Supercomputing Applications (NCSA) on 1–3 March 1995, in Dublin, Ohio, USA, to address the issue of the search and retrieval of data from the internet. The attendees were librarians, archivists, humanities scholars and geographers according to the report from the meeting, along with with IT standardisation experts. This original work was expanded and adopted by various bodies and is now also an ISO standard ISO 15836-1:2017, which establishes 15 core metadata elements for cross-domain resource description, the Dublin Core metadata element set—Part 2: DCMI Properties and Classes is awaiting approval before publication as ISO/DIS 15836-2 due in 2019.
- Subject: The topic addressed by the work
- Title: The name of the object
- Author: The person(s) primarily responsible for the intellectual content of the object
- Publisher: The agent or agency responsible for making the object available
- OtherAgent: The person(s), such as editors and transcribers, who have made other significant intellectual contributions to the work
- Date: The date of publication
- ObjectType: The genre of the object, such as novel, poem, or dictionary
- Form: The physical manifestation of the object, such as Postscript file or Windows executable file
- Identifier: String or number used to uniquely identify the object
- Relation: Relationship to other objects
- Source: Objects, either print or electronic, from which this object is derived, if applicable
- Language: Language of the intellectual content
- Coverage: The spatial locations and temporal durations characteristic of the object
Figure 1. The original 13 Dublin Core Metadata Element Set.
This would lend itself to our spectroscopic data storage system and if necessary we could use the original “Form”, now “Format”, element to indicate the record was a particular IUPAC JCAMP-DX spectroscopy data file, but there is little in here to help or future researcher locate records which meet the search question identified above.
As this discussion continues it is well worth noting a few key observations from the original workshop which we should not lose sight of…
“…indexes are most useful in small collections within a given domain. As the scope of their coverage expands, indexes succumb to problems of large retrieval sets and problems of cross disciplinary semantic drift…”
Or in layman’s terms, what might be stored under the label PULSE SEQUENCE from an NMR spectrum or FID would cause a medical practitioner quite a headache. So, clearly a need exists to separate the technical metadata, which effectively points you towards a specific record on a particular system and will clearly change over time, from the business metadata which gives meaning to the record within a particular discipline or environment. This brings us back to the current question of how to meet the demands of Open Access storage of scientific data in a way which fulfils the FAIR requirements. Fortunately, there is a clear route to managing what the original Dublin Core authors described as the problems of cross disciplinary semantic drift.
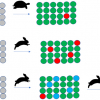
Under the REUSABLE part of the FAIR principles is R1.3. (Meta)data meet domain-relevant community standards. So as a specific data record may be relevant and shared across different “communities” it becomes essential that the metadata which makes a specific data record relevant to that community is clearly separated from what could be an identical metadata term standardised by a different community. Both sets of metadata items can be equally relevant at the time they are generated and have different lifetimes depending on usage (Figure 2).
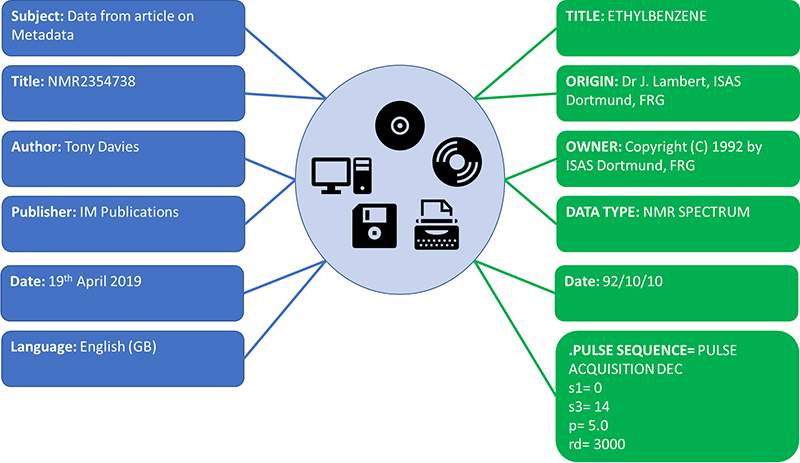
Figure 2. All relevant metadata about the same record but show clear conflicts if the domain relevance of the specific metadata item is not retained.
The GoFair initiative has a nice section explaining what “R1.3. (Meta)data meet domain-relevant community standards” means in practice, which opens the way to integrate our well-established spectroscopic data standards into this environment:
“It is easier to reuse data sets if they are similar: same type of data, data organised in a standardised way, well-established and sustainable file formats, documentation (metadata) following a common template and using common vocabulary. If community standards or best practices for data archiving and sharing exist, they should be followed. For instance, many communities have minimal information standards (e.g., MIAME, MIAPE). FAIR data should at least meet those standards.”
Metadata in regulatory compliance
And this is where this debate gets a lot more serious and is not just some sort of fancy theoretical exercise. As many of you may have read, the Food and Drug Administration has been cracking down on data integrity in the pharmaceutical industry. So much so that they decided to issue a new guidance note in December 2018 in the form of a Question and Answer session to support corporate data compliance in companies. This guidance note helps users understand some of the key underlying regulatory requirement laid out in the so-called predicate rules… in this case those that make up current good manufacturing practice (CGMP) for drugs, as required in 21 CFR parts 210, 211 and 212.5 In case anyone thought that the retention of metadata was not a key component of an overarching data integrity policy the guidance is very clear:
“What is ‘metadata’?
Metadata is the contextual information required to understand data. A data value is by itself meaningless without additional information about the data. Metadata is often described as data about data. Metadata is structured information that describes, explains, or otherwise makes it easier to retrieve, use, or manage data. For example, the number ‘23’ is meaningless without metadata, such as an indication of the unit ‘mg’. Among other things, metadata for a particular piece of data could include a date/time stamp documenting when the data were acquired, a user ID of the person who conducted the test or analysis that generated the data, the instrument ID used to acquire the data, material status data, the material identification number, and audit trails.
Data should be maintained throughout the record’s retention period with all associated metadata required to reconstruct the CGMP activity (e.g., §§ 211.188 and 211.194). The relationships between data and their metadata should be preserved in a secure and traceable manner.”
Of course, none of this is new, but the increased focus on data integrity is now shining a bright spotlight on industry practises and the software solutions we have in place to generate, process, archive and restore our data.
There are two other pieces of guidance in the document which I want to reproduce before we end this article as food for thought...
“9. Can electronic copies be used as accurate reproductions of paper or electronic records?
Yes. Electronic copies can be used as true copies of paper or electronic records, provided the copies preserve the content and meaning of the original record, which includes all metadata required to reconstruct the CGMP activity and the static or dynamic nature of the original records.”
And in question 10 the equivalency of paper and electronic records is discussed, for printouts from pH meters and balances this might satisfy the record retention requirements however,
“10. Is it acceptable to retain paper printouts or static records instead of original electronic records from stand-alone computerized laboratory instruments, such as an FT-IR instrument?
… However, electronic records from certain types of laboratory instruments—whether stand-alone or networked—are dynamic, and a printout or a static record does not preserve the dynamic record format that is part of the complete original record. For example, the spectral file created by FT-IR (Fourier transform infrared) spectroscopy is dynamic and can be reprocessed. However, a static record or printout is fixed and would not satisfy CGMP requirements to retain original records or true copies [§ 211.180(d)]. Also, if the full spectrum is not displayed in the printout, contaminants may be excluded.”
But please go and read the full guidance for all this information to be put into context.
Conclusions
Well for me this whole experience has been a bit of an eye opener. The challenges of getting the data exchange between vendors through a vendor-neutral standardised human-readable format has always been around the data content section and carrying enough metadata through the migrations to ensure the data could be correctly read and interpreted in a second data system (meeting the new FDA guidance explanation of electronic copies needing to preserve the content and meaning of the original record, which includes all metadata required to reconstruct the CGMP activity). So, our new challenge for 2019—to be discussed at the IUPAC 100-year celebrations at the 50th IUPAC General Assembly taking place from 5 to 12 July 2019, in Paris, France—will be to decide what improvements we need to make, together with the instrument vendors, to meet these new metadata challenges. Any volunteers out there?
References
1. L. McEwen, D. Martinsen, R. Lancashire, P. Lampen and A.N. Davies, “Are your spectroscopic data FAIR?”, Spectrosc. Europe 30(4), 21–24 (2018). https://www.spectroscopyeurope.com/td-column/are-your-spectroscopic-data-fair
2. A. Conan Doyle, A Study in Scarlet. Ward Lock (1887).
3. Go FAIR, FAIR Principles. https://www.go-fair.org/fair-principles/
4. http://www.dublincore.org/specifications/dublin-core
5. Data Integrity and Compliance with Drug CGMP, Questions and Answers Guidance for Industry (December 2018). https://www.fda.gov/downloads/drugs/guidances/ucm495891.pdf