Researchers from Ohio State University, USA, have developed a rapid, multiplexed genotyping method to identify the single nucleotide polymorphisms (SNPs) that affect warfarin dose. Their work is reported in the Journal of Molecular Diagnostics(doi: 10.2353/jmoldx.2010.090084).
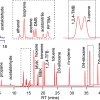
Warfarin is an anti-coagulant that is commonly used to prevent blood clots and embolism. However, warfarin dosing is complicated by the fact that it interacts with many commonly used medications and even chemicals in some foods. Certain genetic variations, SNPs, also affect warfarin sensitivity and metabolism. A group led by Dr Haifeng M. Wu of Ohio StateUniversity has developed a new rapid method to genotype SNPs that will help clinicians to choose appropriate doses of warfarin for individual patients. Using surface-enhanced laser desorption and ionisation time-of-flight mass spectrometry (SELDI-ToF MS), Yang et al. could determine the genotype of three warfarin-related SNPs in under five hours with high levels of accuracy.
Yang et al. suggest that "on-site application of this method in hospital laboratories will greatly help clinicians to determine appropriate doses of warfarin to treat patients with thromboembolic disorders." In future studies, Dr Wu and colleagues plan to apply the SELDI-ToF platform to genotype other medically important SNPs that influence the efficacy and safety profiles of many drug therapies and ultimately to promote personalised health care at Ohio State University.